NASA Earthdata Access in the Cloud Using Open-source libraries
Amy Steiker, NASA National Snow and Ice Data Center DAAC

Contributing co-authors
Catalina M Oaida; NASA PO.DAAC, NASA JPL
Luis Alberto Lopez; NASA National Snow and Ice Data Center DAAC
Aaron Friesz; NASA Land Processes DAAC
Andrew P Barrett; NASA National Snow and Ice Data Center DAAC
Makhan Virdi; NASA ASDC DAAC
Jack McNelis; NASA PO.DAAC, NASA JPL
Julia Lowndes; Openscapes, NCEAS
Erin Robinson; Openscapes, Metadata Game Changers
Additional thanks to the entire NASA Earthdata Openscapes community, Patrick Quinn at Element84, and to 2i2c for our Cloud infrastructure.
Tutorial Outline
- NASA Earthdata discovery and access in the cloud
- Part 1: Explore Earthdata cloud data availablity
- Part 2: Working with Cloud-Optimized GeoTIFFs using NASA’s CMR-STAC
- Part 3: Working with Zarr-formatted data using NASA’s Harmony
- NASA Earthdata’s move to the cloud
- Enabling Open Science via “Analysis-in-Place”
- Resources for cloud adopters: NASA Earthdata Openscapes
- These slides: github.com/NASA-Openscapes/slides
- Cloud Hackathon book: nasa-openscapes.github.io/2021-Cloud-Hackathon
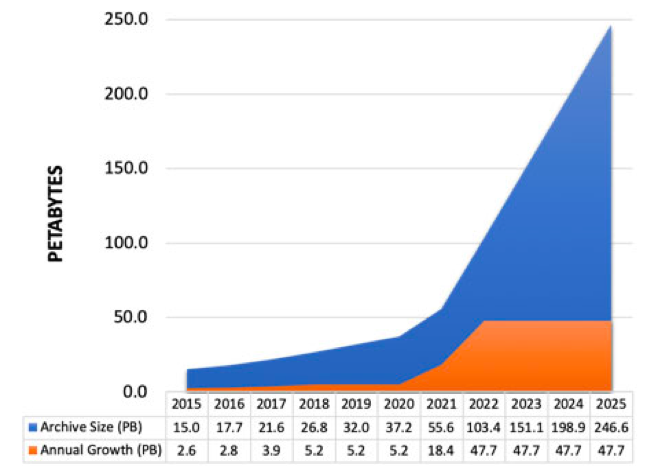
NASA Earthdata archive growth

The NASA Earthdata Cloud Evolution
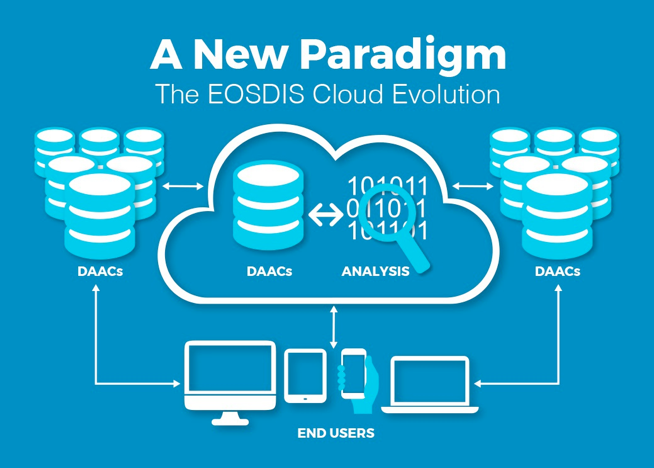
NASA Distributed Active Archive Centers (DAACs) are continuing to migrate data to the Earthdata Cloud
- Supporting increased data volume as new, high-resolution remote sensing missions launch in the coming years
- Data hosted via Amazon Web Services, or AWS
- DAACs continuing to support tools, services, and tutorial resources for our user communities
NASA Earthdata Cloud: Discovery and access w/ open source tech
The following tutorial demonstrates several basic end-to-end workflows to interact with data “in-place” from the NASA Earthdata Cloud, accessing Amazon Web Services (AWS) Single Storage Solution (S3) data locations without the need to download data. While the data can be downloaded locally, the cloud offers the ability to scale compute resources to perform analyses over large areas and time spans, which is critical as data volumes continue to grow.
Although the examples we’re working with in this notebook only focuses on a small time and area for demonstration purposes, this workflow can be modified and scaled up to suit a larger time range and region of interest.
Datasets of interest:
- Harmonized Landsat Sentinel-2 (HLS) Operational Land Imager Surface Reflectance and TOA Brightness Daily Global 30m v2.0 (L30) (10.5067/HLS/HLSL30.002)
- Surface reflectance (SR) and top of atmosphere (TOA) brightness data
- Global observations of the land every 2–3 days at 30-meter (m)
- Cloud Optimized GeoTIFF (COG) format
- ECCO Sea Surface Height - Daily Mean 0.5 Degree (Version 4 Release 4)(10.5067/ECG5D-SSH44).
- Daily-averaged dynamic sea surface height
- Time series of monthly NetCDFs on a 0.5-degree latitude/longitude grid.
Part 1: Explore Earthdata Cloud
Earthdata Search Demo
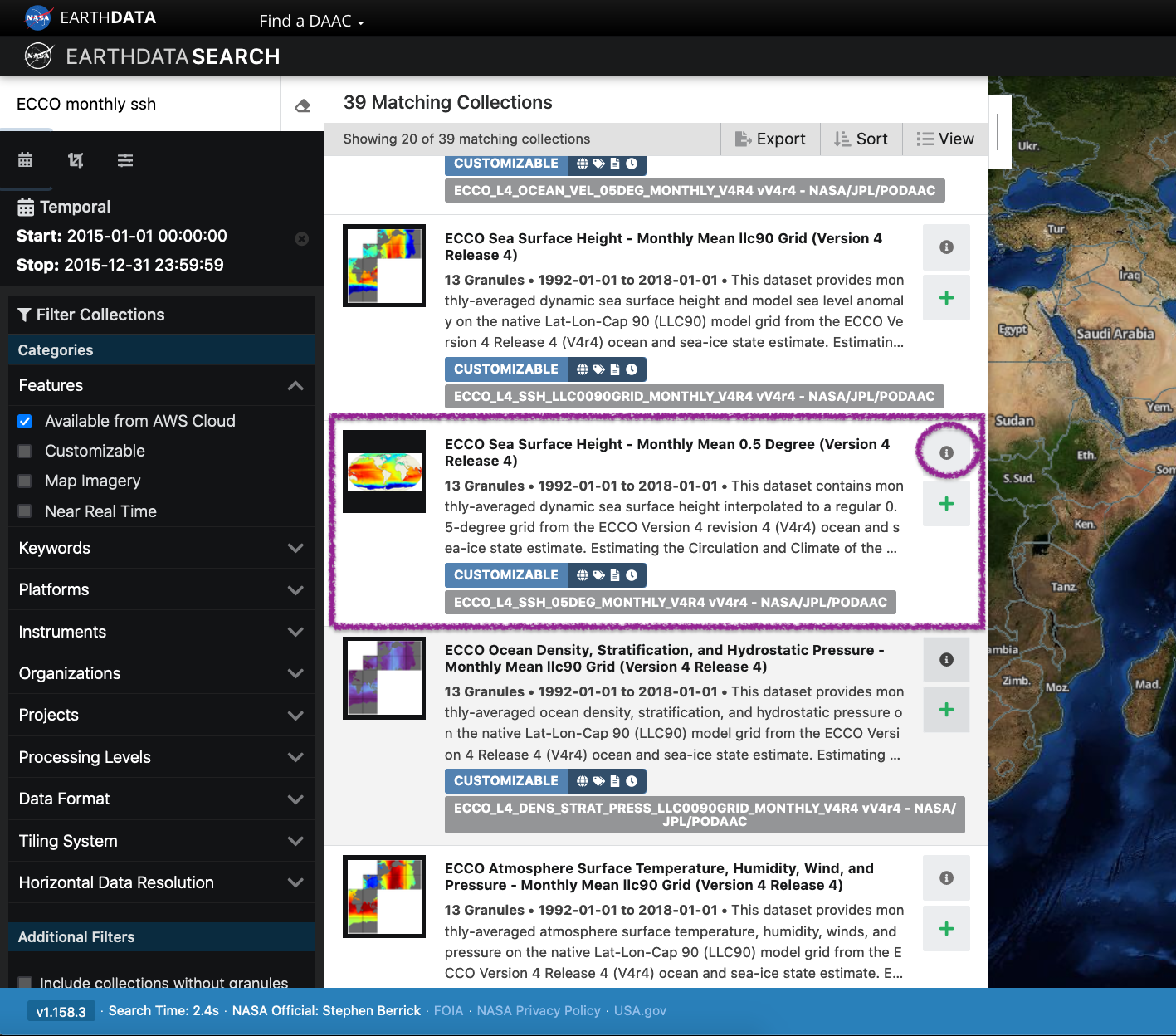
The “Available from AWS Cloud” filter option returns all data from the NASA Earthdata Cloud, including the ECCO dataset, hosted by the PO.DAAC. Here, we search for ECCO monthly SSH over the time period for the year 2015.
View and Select Data Access Options
Clicking on the ECCO Sea Surface Height - Monthly Mean 0.5 Degree (Version 4 Release 4) dataset provides a list of files (granules) that are part of the dataset (collection). There we can select files to add to our project, with options to customize our download or access link(s).
Earthdata Search: Access Options
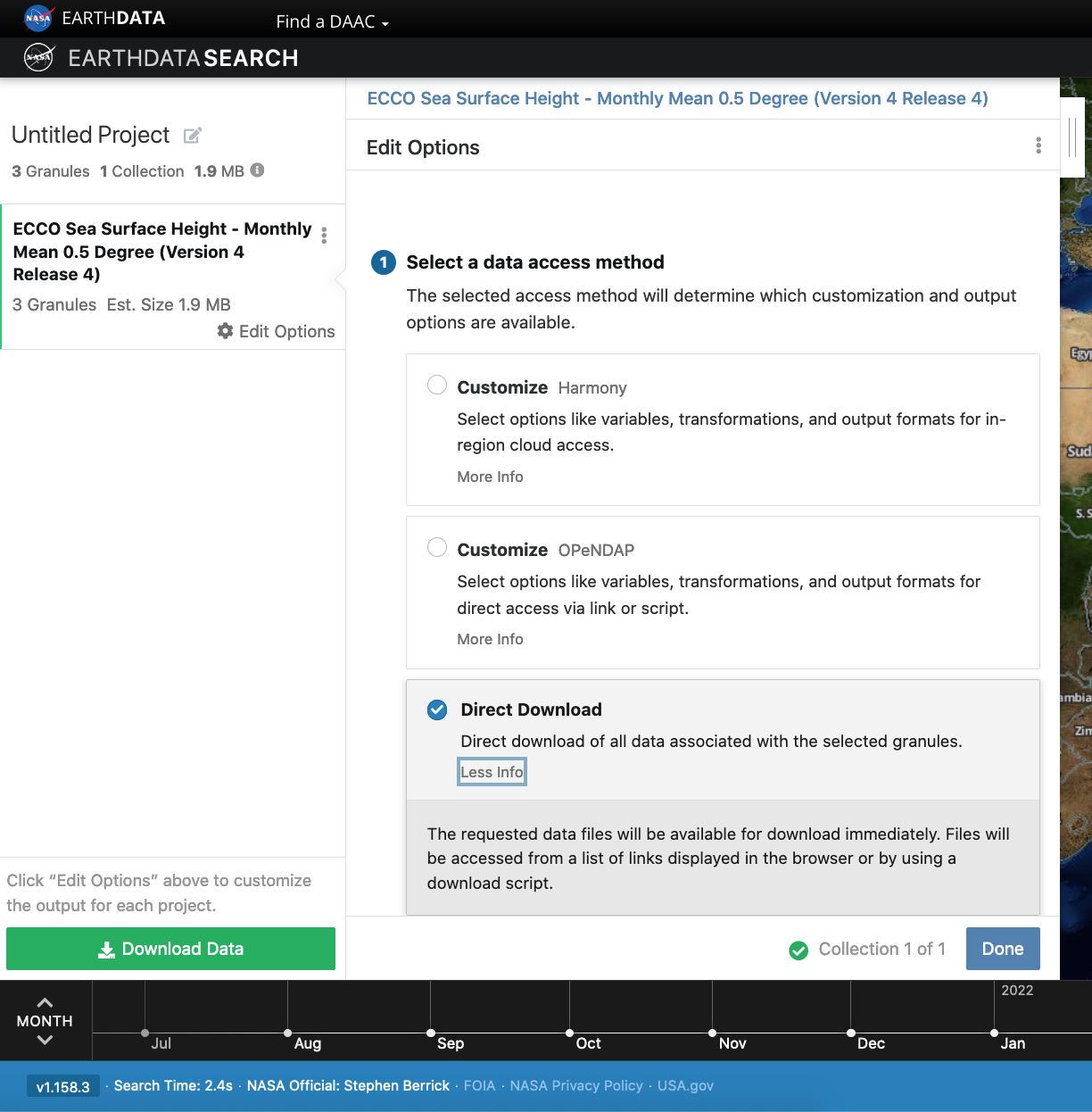
Select the “Direct Download” option to view Access options via Direct Download and from the AWS Cloud. Additional options to customize the data are also available for this dataset.
Earthdata Cloud access information
The final ordering page provides instructions to download and links for data access in the cloud. The AWS S3 Access tab provides the S3:// links, which is what we would use to access the data directly in-region (us-west-2) within the AWS cloud. E.g.: s3://podaac-ops-cumulus-protected/ECCO_L4_SSH_05DEG_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-09_ECCO_V4r4_latlon_0p50deg.nc where s3 indicates data is stored in AWS S3 storage, podaac-ops-cumulus-protected is the bucket, and ECCO_L4_SSH_05DEG_MONTHLY_V4R4 is the object prefix (the latter two are also listed in the dataset collection information under Cloud Access (step 3 above)).
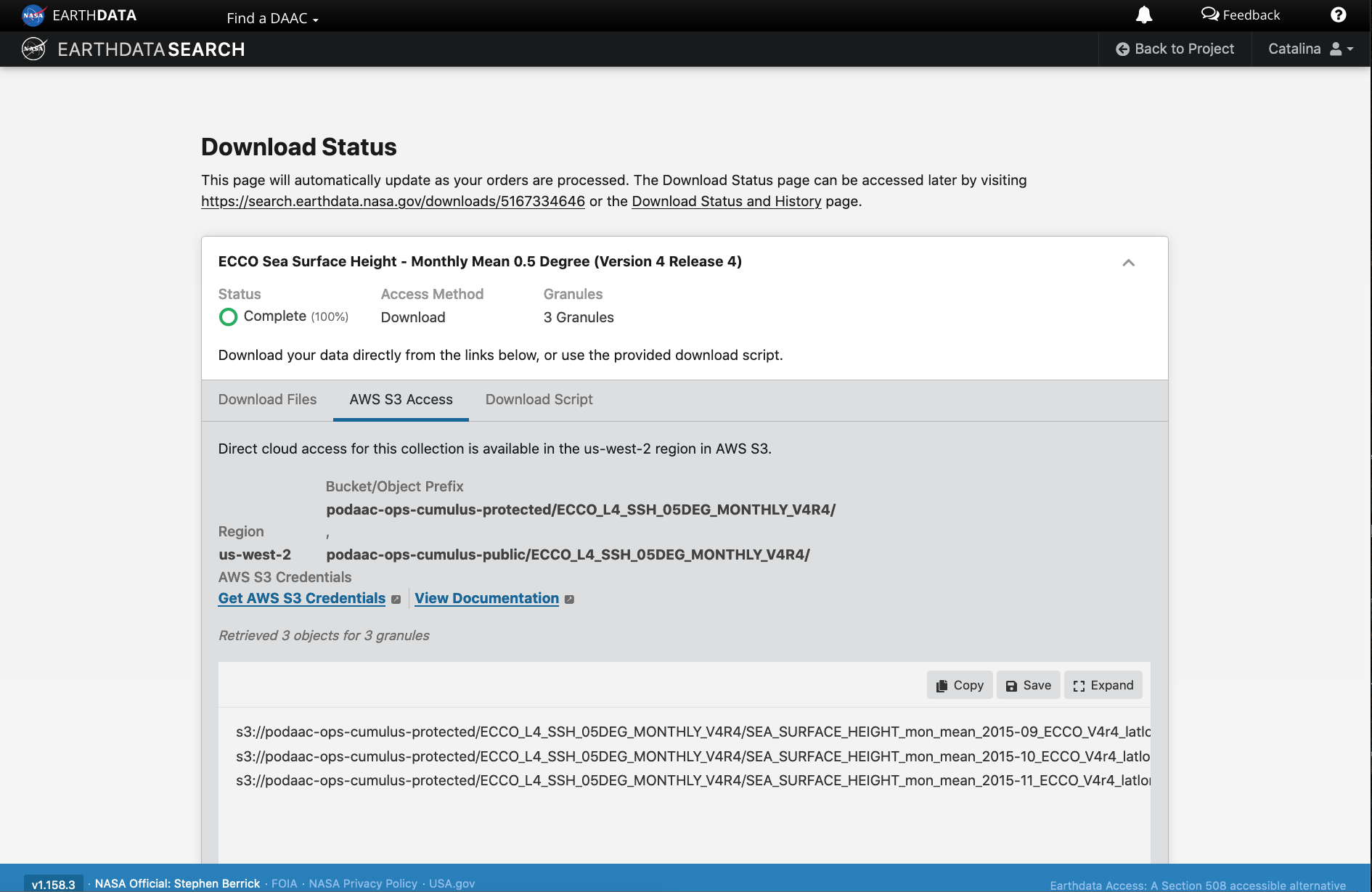
Earthdata Cloud access information

Integrate file links into programmatic workflow, locally or in the AWS cloud.
We can connect these access links to subsequent data analysis in the cloud by either copy/pasting the s3:// links or saving them as a text file to then access in a Jupyter notebook or script running in the cloud.
Part 2: Working with Cloud-Optimized GeoTIFFs
using NASA’s Common Metadata Repository SpatioTemporal Assett Catalog (CMR-STAC)
In this example we will access the NASA’s Harmonized Landsat Sentinel-2 (HLS) version 2 assets, which are archived in cloud optimized geoTIFF (COG) format archived by the Land Processes (LP) DAAC. The COGs can be used like any other GeoTIFF file, but have some added features that make them more efficient within the cloud data access paradigm. These features include: overviews and internal tiling.
But first, what is STAC?
SpatioTemporal Asset Catalog (STAC) is a specification that provides a common language for interpreting geospatial information in order to standardize indexing and discovering data.
The STAC specification is made up of a collection of related, yet independent specifications that when used together provide search and discovery capabilities for remote assets.
| Four STAC Specifications: | |
|---|---|
| STAC Catalog (aka DAAC Archive) | STAC Collection (aka Data Product) |
| STAC Item (aka Granule) | STAC API |
CMR-STAC API
The CMR-STAC API is NASA’s implementation of the STAC API specification for all NASA data holdings within EOSDIS. The current implementation does not allow for querries accross the entire NASA catalog. Users must execute searches within provider catalogs (e.g., LPCLOUD) to find the STAC Items they are searching for. All the providers can be found at the CMR-STAC endpoint here: https://cmr.earthdata.nasa.gov/stac/.
In this example, we will query the LPCLOUD provider to identify STAC Items from the Harmonized Landsat Sentinel-2 (HLS) collection that fall within our region of interest (ROI) and within our specified time range.
Connect to the CMR-STAC API:
And the Land Processes DAAC LPCLOUD Provider/STAC Catalog:
For this next step we need the provider title (e.g., LPCLOUD). We will add the provider to the end of the CMR-STAC API URL (i.e., https://cmr.earthdata.nasa.gov/stac/) to connect to the LPCLOUD STAC Catalog.
Since we are using a dedicated client (i.e., pystac-client.Client) to connect to our STAC Provider Catalog, we will have access to some useful internal methods and functions (e.g., get_children() or get_all_items()) we can use to get information from these objects.
Search for STAC Items: Read in a geojson file and plot
We will define our ROI using a geojson file containing a small polygon feature in western Nebraska, USA. We’ll also specify the data collections and a time range for our example.
Read in a geojson file with geopandas and extract coodinates for our ROI. We can plot the polygon using the geoviews package that we imported as gv with ‘bokeh’ and ‘matplotlib’ extensions. The following has reasonable width, height, color, and line widths to view our polygon when it is overlayed on a base tile map.
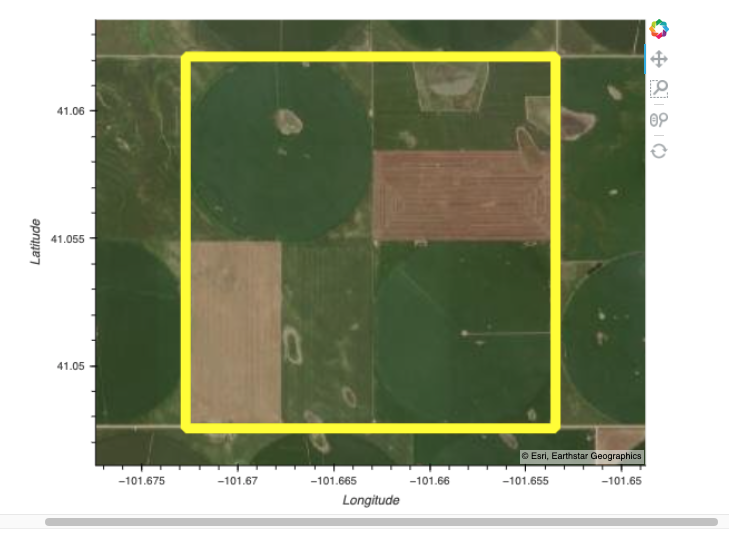
Search the CMR-STAC API with our search criteria
Now we can put all our search criteria together using catalog.search from the pystac_client package. STAC Collection is synonomous with what we usually consider a NASA data product. Desired STAC Collections are submitted to the search API as a list containing the collection id. Let’s focus on S30 and L30 collections.
View STAC Items that matched our search query
print('Matching STAC Items:', search.matched())
item_collection = search.get_all_items()
item_collection[0].to_dict()Matching STAC Items: 113{'type': 'Feature',
'stac_version': '1.0.0',
'id': 'HLS.L30.T13TGF.2021124T173013.v2.0',
'properties': {'datetime': '2021-05-04T17:30:13.428000Z',
'start_datetime': '2021-05-04T17:30:13.428Z',
'end_datetime': '2021-05-04T17:30:37.319Z',
'eo:cloud_cover': 36},
'geometry': {'type': 'Polygon',
'coordinates': [[[-101.5423534, 40.5109845],
[-101.3056118, 41.2066375],
[-101.2894253, 41.4919436],
[-102.6032964, 41.5268623],
[-102.638891, 40.5386175],
[-101.5423534, 40.5109845]]]},
'links': [{'rel': 'self',
'href': 'https://cmr.earthdata.nasa.gov/stac/LPCLOUD/collections/HLSL30.v2.0/items/HLS.L30.T13TGF.2021124T173013.v2.0'},
{'rel': 'parent',
'href': 'https://cmr.earthdata.nasa.gov/stac/LPCLOUD/collections/HLSL30.v2.0'},
{'rel': 'collection',
'href': 'https://cmr.earthdata.nasa.gov/stac/LPCLOUD/collections/HLSL30.v2.0'},
{'rel': <RelType.ROOT: 'root'>,
'href': 'https://cmr.earthdata.nasa.gov/stac/LPCLOUD/',
'type': <MediaType.JSON: 'application/json'>,
'title': 'LPCLOUD'},
{'rel': 'provider', 'href': 'https://cmr.earthdata.nasa.gov/stac/LPCLOUD'},
{'rel': 'via',
'href': 'https://cmr.earthdata.nasa.gov/search/concepts/G2144020713-LPCLOUD.json'},
{'rel': 'via',
'href': 'https://cmr.earthdata.nasa.gov/search/concepts/G2144020713-LPCLOUD.umm_json'}],
'assets': {'B11': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B11.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B11.tif'},
'B07': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B07.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B07.tif'},
'SAA': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.SAA.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.SAA.tif'},
'B06': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B06.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B06.tif'},
'B09': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B09.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B09.tif'},
'B10': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B10.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B10.tif'},
'VZA': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.VZA.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.VZA.tif'},
'SZA': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.SZA.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.SZA.tif'},
'B01': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B01.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B01.tif'},
'VAA': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.VAA.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.VAA.tif'},
'B05': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B05.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B05.tif'},
'B02': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B02.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B02.tif'},
'Fmask': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.Fmask.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.Fmask.tif'},
'B03': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B03.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B03.tif'},
'B04': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.B04.tif',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.B04.tif'},
'browse': {'href': 'https://data.lpdaac.earthdatacloud.nasa.gov/lp-prod-public/HLSL30.020/HLS.L30.T13TGF.2021124T173013.v2.0/HLS.L30.T13TGF.2021124T173013.v2.0.jpg',
'type': 'image/jpeg',
'title': 'Download HLS.L30.T13TGF.2021124T173013.v2.0.jpg'},
'metadata': {'href': 'https://cmr.earthdata.nasa.gov/search/concepts/G2144020713-LPCLOUD.xml',
'type': 'application/xml'}},
'bbox': [-102.638891, 40.510984, -101.289425, 41.526862],
'stac_extensions': ['https://stac-extensions.github.io/eo/v1.0.0/schema.json'],
'collection': 'HLSL30.v2.0'}Filtering STAC Items
Below we will loop through and filter the item_collection by a specified cloud cover as well as extract the band we’d need to do an Enhanced Vegetation Index (EVI) calculation for a future analysis. We will also specify the STAC Assets (i.e., bands/layers) of interest for both the S30 and L30 collections (also in our collections variable above) and print out the first ten links, converted to s3 locations:
cloudcover = 25
s30_bands = ['B8A', 'B04', 'B02', 'Fmask'] # S30 bands for EVI calculation and quality filtering -> NIR, RED, BLUE, Quality
l30_bands = ['B05', 'B04', 'B02', 'Fmask'] # L30 bands for EVI calculation and quality filtering -> NIR, RED, BLUE, Quality
evi_band_links = []
for i in item_collection:
if i.properties['eo:cloud_cover'] <= cloudcover:
if i.collection_id == 'HLSS30.v2.0':
#print(i.properties['eo:cloud_cover'])
evi_bands = s30_bands
elif i.collection_id == 'HLSL30.v2.0':
#print(i.properties['eo:cloud_cover'])
evi_bands = l30_bands
for a in i.assets:
if any(b==a for b in evi_bands):
evi_band_links.append(i.assets[a].href)
s3_links = [l.replace('https://data.lpdaac.earthdatacloud.nasa.gov/', 's3://') for l in evi_band_links]
s3_links[:10]Filtering STAC Items
Below we will loop through and filter the item_collection by a specified cloud cover as well as extract the band we’d need to do an Enhanced Vegetation Index (EVI) calculation for a future analysis. We will also specify the STAC Assets (i.e., bands/layers) of interest for both the S30 and L30 collections (also in our collections variable above) and print out the first ten links, converted to s3 locations:
['s3://lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021133T172406.v2.0/HLS.L30.T13TGF.2021133T172406.v2.0.B04.tif',
's3://lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021133T172406.v2.0/HLS.L30.T13TGF.2021133T172406.v2.0.B05.tif',
's3://lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021133T172406.v2.0/HLS.L30.T13TGF.2021133T172406.v2.0.Fmask.tif',
's3://lp-prod-protected/HLSL30.020/HLS.L30.T13TGF.2021133T172406.v2.0/HLS.L30.T13TGF.2021133T172406.v2.0.B02.tif',
's3://lp-prod-protected/HLSL30.020/HLS.L30.T14TKL.2021133T172406.v2.0/HLS.L30.T14TKL.2021133T172406.v2.0.B02.tif',
's3://lp-prod-protected/HLSL30.020/HLS.L30.T14TKL.2021133T172406.v2.0/HLS.L30.T14TKL.2021133T172406.v2.0.B04.tif',
's3://lp-prod-protected/HLSL30.020/HLS.L30.T14TKL.2021133T172406.v2.0/HLS.L30.T14TKL.2021133T172406.v2.0.B05.tif',
's3://lp-prod-protected/HLSL30.020/HLS.L30.T14TKL.2021133T172406.v2.0/HLS.L30.T14TKL.2021133T172406.v2.0.Fmask.tif',
's3://lp-prod-protected/HLSS30.020/HLS.S30.T14TKL.2021133T173859.v2.0/HLS.S30.T14TKL.2021133T173859.v2.0.B04.tif',
's3://lp-prod-protected/HLSS30.020/HLS.S30.T14TKL.2021133T173859.v2.0/HLS.S30.T14TKL.2021133T173859.v2.0.B8A.tif']Access s3 storage location
Access s3 credentials from LP.DAAC and create a boto3 Session object using your temporary credentials. This Session is used to pass credentials and configuration to AWS so we can interact wit S3 objects from applicable buckets.
s3_cred_endpoint = 'https://data.lpdaac.earthdatacloud.nasa.gov/s3credentials'
temp_creds_req = requests.get(s3_cred_endpoint).json()
session = boto3.Session(aws_access_key_id=temp_creds_req['accessKeyId'],
aws_secret_access_key=temp_creds_req['secretAccessKey'],
aws_session_token=temp_creds_req['sessionToken'],
region_name='us-west-2')GDAL Configurations
GDAL is a foundational piece of geospatial software that is leveraged by several popular open-source, and closed, geospatial software. The rasterio package is no exception. Rasterio leverages GDAL to, among other things, read and write raster data files, e.g., GeoTIFFs/Cloud Optimized GeoTIFFs. To read remote files, i.e., files/objects stored in the cloud, GDAL uses its Virtual File System API. In a perfect world, one would be able to point a Virtual File System (there are several) at a remote data asset and have the asset retrieved, but that is not always the case. GDAL has a host of configurations/environmental variables that adjust its behavior to, for example, make a request more performant or to pass AWS credentials to the distribution system. Below, we’ll identify the evironmental variables that will help us get our data from cloud.
Read Cloud-Optimized GeoTIFF into rioxarray
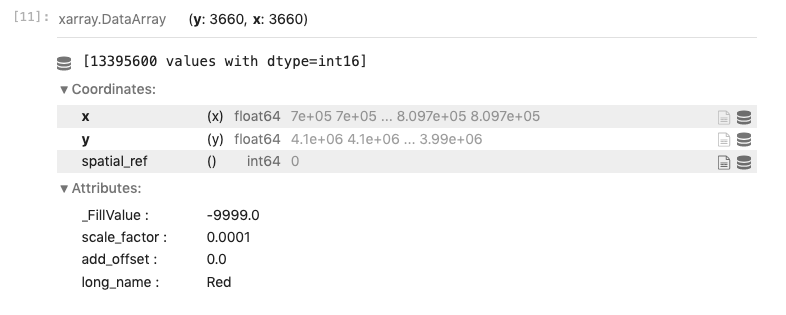
Plot using hvplot

Part 3: Working with Zarr-formatted data
using NASA’s Harmony cloud transformation service
We have already explored direct access to the NASA EOSDIS archive in the cloud via the AWS S3. In addition to directly accessing the files archived and distributed by each of the NASA DAACs, many datasets also support services that allow us to customize the data via subsetting, reformatting, reprojection, and other transformations.
This example demonstrates “analysis in place” using customized ECCO Level 4 monthly sea surface height data, in this case reformatted to Zarr, from a new ecosystem of services operating within the NASA Earthdata Cloud: NASA Harmony:
- Consistent access patterns to EOSDIS holdings make cross-data center data access easier
- Data reduction services allow us to request only the data we want, in the format and projection we want
- Analysis Ready Data and cloud access will help reduce time-to-science
- Community Development helps reduce the barriers for re-use of code and sharing of domain knowledge
Using Harmony-Py to customize data
Harmony-Py provides a pip installable Python alternative to directly using Harmony’s OGC Coverages API to make it easier to request data and service options, especially when interacting within a Python Jupyter Notebook environment.
Create Harmony Client object
First, we need to create a Harmony Client, which is what we will interact with to submit and inspect a data request to Harmony, as well as to retrieve results.
Create Harmony Request
Specify a temporal range over 2015, and Zarr as an output format.
What is Zarr?
Zarr is an open source library for storing N-dimensional array data. It supports multidimensional arrays with attributes and dimensions similar to NetCDF4, and it can be read by XArray. Zarr is often used for data held in cloud object storage (like Amazon S3), because it is better optimized for these situations than NetCDF4.
Check request status and view output URLs
Harmony data outputs can be accessed within the cloud using the s3 URLs and AWS credentials provided in the Harmony job response:
harmony_client.wait_for_processing(job_id, show_progress=True)
results = harmony_client.result_urls(job_id, link_type=LinkType.s3)
s3_urls = list(results)
s3_urls['s3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-01_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-02_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-12_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-11_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-03_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-04_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-06_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-05_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-07_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-08_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-09_ECCO_V4r4_latlon_0p50deg.zarr',
's3://harmony-prod-staging/public/harmony/netcdf-to-zarr/0bfeca8d-8aa7-4dc0-920c-92491d91dd55/SEA_SURFACE_HEIGHT_mon_mean_2015-10_ECCO_V4r4_latlon_0p50deg.zarr']Open staged files with s3fs and xarray
Access AWS credentials for the Harmony bucket, and use the AWS s3fs package to create a file system that can then be read by xarray. Below we create session by passing in the temporary credentials we recieved from our temporary credentials endpoint.
Open staged files with s3fs and xarray
Open the Zarr stores using the s3fs package, then load them all at once into a concatenated xarray dataset:
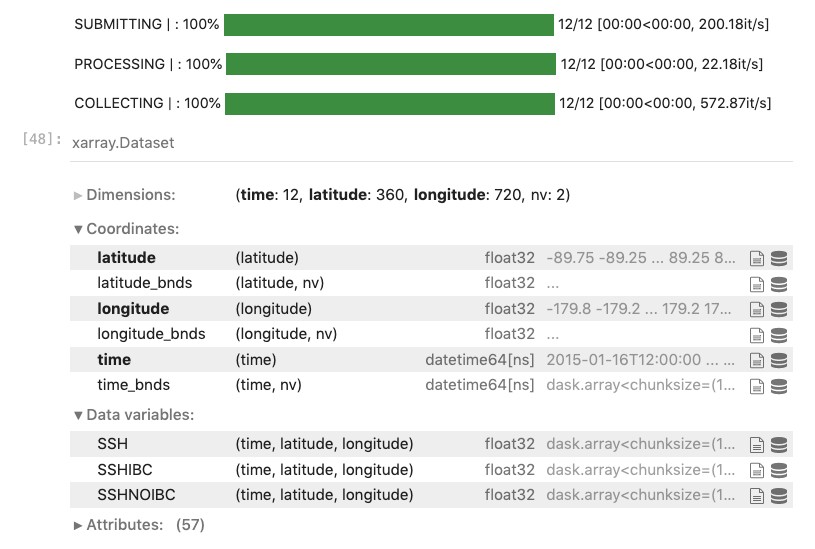
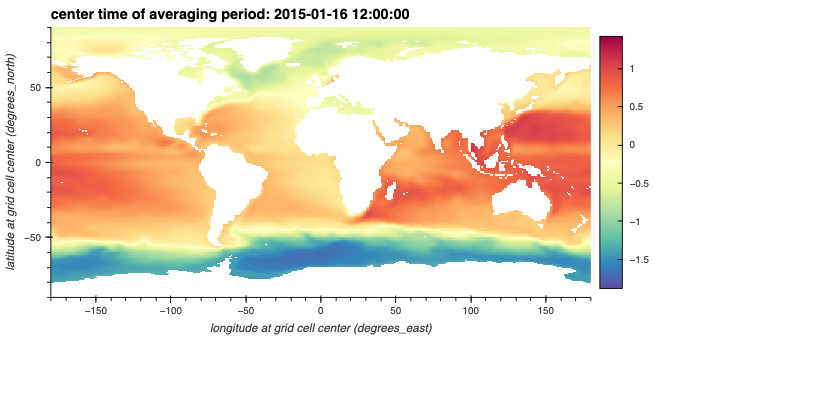
Plot the Sea Surface Height time series using hvplot
Now we can start looking at aggregations across the time dimension. Here we plot the SSH variable using hvplot and can use the time slider to visualize changes in SSH over the year.
Data and Analysis co-located “in place”
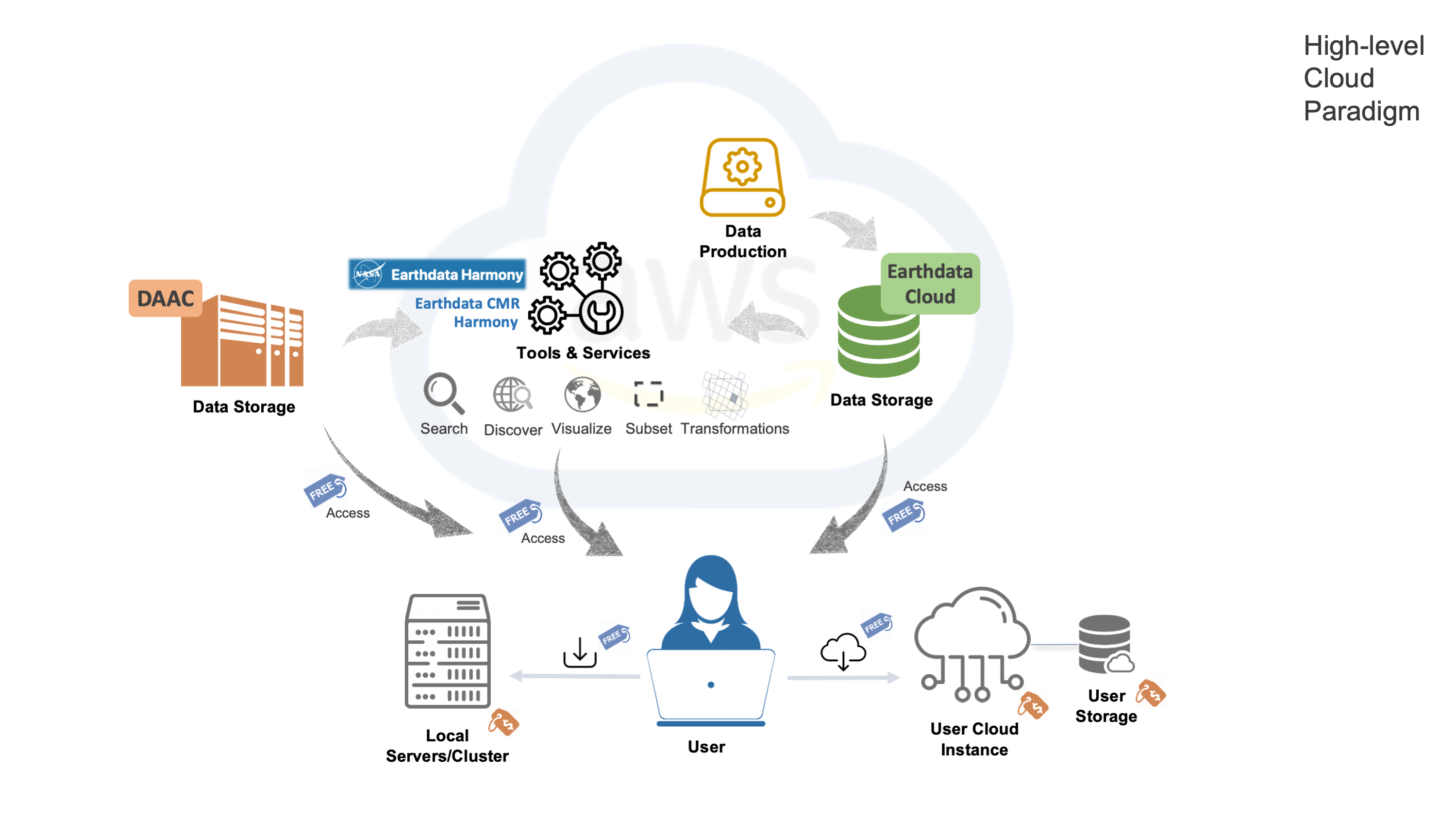
NASA Earthdata Cloud as an enabler of Open Science

*Reducing barriers to large-scale scientific research in the era of “big data”
*Increasing community contributions with hands-on engagement
*Promoting reproducible and shareable workflows without relying on local storage systems
Building NASA Earthdata Cloud Resources

A Growing List of Resources!
NASA Earthdata Cloud Primer -AWS cloud primer: helpful tutorials for how to set up your own EC2 cloud instance in AWS, attach storeage, move files back and forth, and more.
Setting up a JupyterHub on AWS - Earthdata Cloud Hackathon participant’s repo replicating our shared compute environment to access and analyze Earthdata “in place”
USGS Eyes on Earth Podcast: Satellites and Cloud Computing - with Aaron Friesz (LP DAAC!)